Interesting Paper: Land-Sparing Agriculture Best Protects Avian Phylogenetic Diversity
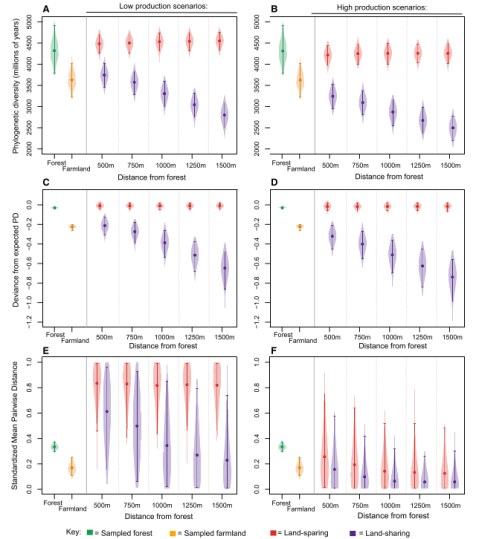
A quick post to highlight a new publication in this weeks issue of Current Biology. Edwards et al. went for another piece on the land-sharing/land-sparing debate and presented a very nice case study. Land-sharing is often defined as combining “sustainable” agricultural production with higher biodiversity outcomes often at the tradeoff of harvesting less and loss of natural habitats. Land-sparing on the other hand attempts to prevent remaining natural habitat from being used by humans, but instead intensify production and increase yield from other areas, thus reducing their potential for wildlife-friendly farming. They combined field work from the Choco-andres region (Taxonomic focus: Birds) with simulation models to investigate which strategy might benefit biodiversity the most. Contrary to many other previous publications they focused on phylogenetic richness (PD) rather than “species richness”. Based on landscape simulation models they could show that PD decreases steadily with greater distance to forests, which is interesting because it demonstrates that land-sharing strategies might only be successful, if sufficient amounts of natural habitat are in close proximity, that can act as source habitat for dispersing species.
According to their analysis some species seem to benefit more from land-sparing strategies than others. Specific evolutionary traits thus might be ether beneficial or detrimental for surviving in intensive human land use such as agriculture. They conclude that land-sharing might be of limited benefit without the simultaneous protection of nearby blocks of natural habitat, which can only be achieved with a co-occurring land-sharing strategy.
Further reading: http://www.theguardian.com/science/grrlscientist/2015/sep/03/farming-practices-benefit-tropical-birds-agriculture-sustainability
Assessing habitat specialization using IUCN data
Since quite some time ecological models have tried to incorporate both continuous and discrete characteristics of species into their models. Newbold et al. (2013) demonstrated that functional traits affect the response of tropical bird species towards land-use intensity. Tropical forest specialist birds seem to decrease globally in probability of presence and abundance in more intensively used forests. This patterns extends to many taxonomic groups and the worldwide decline of “specialist species” has been noted before by Clavel et al. (2011).
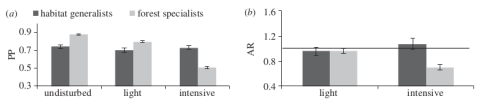
(a) Probabilities of presence of tropical bird species in in different disturbed forests and (b) ratios of abundance in light and intensive disturbed forests relative to undisturbed forests. Forest specialists are disproportionally affected in intensively used forests. Figure from Newbold et al. 2013 doi: http://dx.doi.org/10.1098/rspb.2012.2131
But how to acquire such data on habitat specialization? Ether you assemble your own exhaustive trait database or you query information from some of the openly available data sources. One could for instance be the IUCN redlist, which not only has expert-validated data on a species current threat status, but also on population size and also on a species habitat preference. Here IUCN follows its own habitat classification scheme ( http://www.iucnredlist.org/technical-documents/classification-schemes/habitats-classification-scheme-ver3 ). The curious ecologist and conservationist should keep in mind however, that not all species are currently assessed by IUCN.
There are already a lot of scripts available on the net from which you can get inspiration on how to query the IUCN redlist (Kay Cichini from the biobucket explored this already in 2012 ). Even better: Someone actually compiled a whole r-package called letsR full of web-scraping functions to access the IUCN redlist. Here is some example code for Perrin’s Bushshrike, a tropical bird quite common in central Africa
# Install package install.packages(letsR) library(letsR) # Perrin's or Four-colored Bushshrike latin name name <- 'Telophorus viridis' # Query IUCN status lets.iucn(name) #>Species Family Status Criteria Population Description_Year #>Telophorus viridis MALACONOTIDAE LC Stable 1817 #>Country #>Angola, Congo, The Democratic Republic of the Congo, Gabon, Zambia # Or you can query habitat information lets.iucn.ha(name) #>Species Forest Savanna Shrubland Grassland Wetlands Rocky areas Caves and Subterranean Habitats #>Telophorus viridis 1 1 1 0 0 0 0 #> Desert Marine Neritic Marine Oceanic Marine Deep Ocean Floor Marine Intertidal Marine Coastal/Supratidal #> 0 0 0 0 0 0 #> Artificial/Terrestrial Artificial/Aquatic Introduced Vegetation Other Unknown #> 1 0 0 0 0
letsR also has other methods to work with the spatial data that IUCN provides ( http://www.iucnredlist.org/technical-documents/spatial-data ), so definitely take a look. It works by querying the IUCN redlist api for the species id (http://api.iucnredlist.org/go/Telophorus-viridis). Sadly the habitat function does only return the information if a species is known to occur in a given habitat, but not if it is of major importance for a particular species (so if for instance a Species is known to be a “forest-specialist” ). Telophorus viridis for instance also occurs in savannah and occasionally artificial habitats like gardens ( http://www.iucnredlist.org/details/classify/22707695/0 ).
So I just programmed my own function to assess if forest habitat is of major importance to a given species. It takes a IUCN species id as input and returns ether “Forest-specialist”, if forest habitat is of major importance to a species, “Forest-associated” if a species is just known to occur in forest or “Other Habitats” if a species does not occur in forests at all. The function works be cleverly querying the IUCN redlist and breaking up the HTML structure at given intervals that indicate a new habitat type.
Find the function on gist.github (Strangely WordPress doesn’t include them as they promised)
How does it work? You first enter the species IUCN redlist id. It is in the url after you have queried a given species name. Alternatively you could also download the whole IUCN classification table and match your species name against it 😉 Find it here. Then simply execute the function with the code.
name = 'Telophorus viridis'
data <- read.csv('all.csv')
# This returns the species id
data$Red.List.Species.ID[which(data$Scientific.Name==name)]
#> 22707695
# Then simply run my function
isForestSpecialist(22707695)
#> 'Forest-specialist'
Out in the field – Working in the agricultural Mosaic of the Taita Hills
And here are some news from my current field work that is part of my Thesis. After spending some quiet, but exiting days in Nairobi (maybe later more about that) I finally arrived in Wundanyi, Taita Hills, where a substantial part of my work will be conducted along the CHIESA transect. Suited in the coastel area in proximity to Mombasa the Taita Hills are renown for their extraordinary bird diversity and endemic species and as such are considered to be part of the Eastern Arc Mountains Diversity hotspot. The Taita hills encompass a variety of different land-use forms, but the majority of them surely are tropical homegardens as most of the “Taita” people are subsistence farmers growing crops in the highly fertile soil of the mountain slopes. Besides homegardens there are riverine forests in the valleys, shrubland vegetation in the lower altitudes, exotic tree plantations and of course the remaining indigenous forests remaining on the Taita hills mountain tops. Every last forest part is known well and was traditionally protected by the locals as part of their culture. However in the later centuries the remaining forest area became more and more scarcer and even during my visits in some of the forest fragments with the highest biodiversity value (Chawia, Ngangao) I saw frequent signs of fuelwood and timber extraction. Clearly a lack of funding for biodiversity protection seems to be the problem, but also an economic perspective and opportunities such as ecotourism might enhance locals perception if and how these last forest parts should be protected.
My work in the Taita hills is all about birds. Specifically I am conducting avian diversity and abundance assessments along an altitudinal transect encompassing a variety of different land-use systems. Although avian assessments have been conducted in Taita many times before, they were often restricted to the forest fragments and for instance didn’t look at the bird diversity in homegardens in different altitudes. The resulting data will just be used for my thesis as validation dataset, but I am hoping that it has maybe some value on its own as well. Initial results show that especially the homegarden in Taita support quite a high diversity of birds, which is even similar to levels in the remaining forest fragments (although the community is somewhat different and biotic homogenization is likely on-going).
It can be quite challenging to conduct avian research in tropical human-dominated landscapes. Not only do you have to arrange for transport to the specific transect areas and lodging (in my case provided by the University of Helsinki Research station in Wundanyi), but also account for the frequent interruption by children and farmers asking what you are doing. Furthermore it is not an easy task to count birds in for instance a maize or sugarcane plantation due to the limited accessibility and my intention not to damage the farmers crops. Most of the farmers however happily provide access to their land and are very interested in what kind of research this “Mzungu” is doing on their farm. From my own experience here I can tell that the Taita people are very kind and it is a pleasure to work with them on their land. They are very respectful and even walking around late at night or very early in the morning seems to be no problem here (in contrast to for instance Nairobi or Mombasa).
In the end my sampling goes on quite well and much better than I expected. Although it is technically raining season and long heavy rains can be expected every day, the mornings were exceptionally dry and weather was mostly favourable for ornithological research. Generally this time of the year in East Africa is especially interesting for bird assessments as many local bird species are in their breeding plumage and nesting, but also because European migrants are often still around or on their way back to Europe (for instance I saw and heard an European Willow Warbler some days ago). Lets see what else the next weeks will have for be in terms of avian diversity.
Macroecology playground (1) – Bird species richness in a nutshell
Ahh, Macroecology. The study of ecological patterns and processes on big scales. Questions like “what factors determine distribution and diversity of all life on earth?” have troubled scientists since A.v.Humboldt and Wallace times. At the University of Copenhagen a whole research center has been dedicated to this specific field and macro-ecological studies are more and more present in prestigious journals like Nature and Science. Previous studies at the center have found skewed distributions of bird richness with a specific bias towards the mountains (Jetz & Rahbek, 2002, Rahbek et al., 2007). In this blog post i am going to play a bit around with some data from Rahbek et al. (2007). The analysis and the graphs are by no means sufficient (and even violate many model assumptions like homoscedasticity, normality and data independence) and are therefore more of exploratory nature 😉 The post will show you how to build a raster stack of geographical data and how to use the data in some very basic models.
It was recommended to me to use the freely available SAM software for the analysis but although the program is really nice and fast it isn’t suitable enough for me as you can not modify specific model parameters or graphical outputs. And as a self-declared R junkie i refuse to work with “click-compute-result” tools 😉
So here is how the head of SAM data file (“data.sam”) looks like (i won’t share it, so please generate your own data).
![]() As you can see the .sam file is technically just a tabulator separated table with the coordinates for a gridcell (1° gridcell on a latitude-longitude projection) and all response and predictor values for this cell. To get this data into R we are gonna use the raster package to generate a so called raster stack for our analysis. This is how i did it
As you can see the .sam file is technically just a tabulator separated table with the coordinates for a gridcell (1° gridcell on a latitude-longitude projection) and all response and predictor values for this cell. To get this data into R we are gonna use the raster package to generate a so called raster stack for our analysis. This is how i did it
# Load libraries
library(raster)
# Create Data from SAM
data <- read.delim(file="data.sam",header=T,sep="\t",dec=".") # read in a data.frame
coordinates(data) <- ~Longitude+Latitude # Convert to a SpatialPointsDataframe
cs <- "+proj=longlat +datum=WGS84 +no_defs" # define the correct projection (long-lat)
gridded(data) <- T # Make a SpatialPixelsDataframe
proj4string(data) <- CRS(cs) # set the defined CRS
# Create Raster layer stack
s <- stack()
for(n in names(data)){
d <- data.frame(coordinates(data),data[,n])
ras <- rasterFromXYZ(xyz=d,digits=10,crs=CRS(cs))
s <- addLayer(s,ras)
rm(d,n,ras)
}
# Now you can query and plot the raster layers from the stack
plot(s$Birds.richness,col=rainbow(100,start=0.1))
You wanna do some modeling or extract data? Here you go. First we make a subset of some of our predictors from the raster stack and then fit ordinary least squares multiple regression models to our data to see how much variance can be explained. Note that linear regressions are not the proper techniques for this kind of analysis (degrees of freedom to high due to spatial autocorrelation, violation of assumptions mentioned before), but its still useful for explanatory purposes.
# Extract some predictors from the raster Stack predictors <- subset(s,c(7,8,10)) names(predictors) > "NDVI" "Topographical.Range" "Annual.Mean.Temperature" # Now extract the data from both the bird richness layer and the predictors birds <- getValues(s$Birds.richness) val <- as.data.frame(getValues(predictors)) # Do the multiple regression fit <- lm(birds~.,data=val) summary(fit) > Estimate Std. Error t value Pr(>|t|) (Intercept) 215.675282 15.837493 13.62 <2e-16 *** NDVI -34.541242 1.245769 -27.73 <2e-16 *** Topographical.Range 0.056458 0.002452 23.03 <2e-16 *** Annual.Mean.Temperature 0.940664 0.054747 17.18 <2e-16 *** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 81.86 on 1525 degrees of freedom (1461 observations deleted due to missingness) Multiple R-squared: 0.6931, Adjusted R-squared: 0.6925 F-statistic: 1148 on 3 and 1525 DF, p-value: < 2.2e-16
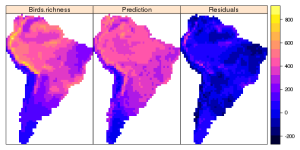
Ignore the p-values and just focus on the adjusted r² value. As you can see we are able to explain nearly 70% of the variance with this simple model. So how do our residuals and the predicted values look like? For that we have to create analogous raster layers containing both the predicted and the residual values. Then we plot all species raster layers again using the spplot function from the package sp (automatically loaded with “raster”)
# Estimates prediction rval <- getValues(s$Birds.richness) # Create new values rval[as.numeric(names(fit$fitted.values))]<- predict(fit) # replace all data-cells with predicted values pred <- predictors$NDVI # make a copy of an existing raster values(pred) <-rval;rm(rval) #replace all values in this raster copy names(pred) <- "Prediction" # Residual Raster rval <- getValues(s$Birds.richness) # Create new values rval[as.numeric(names(fit$residuals))]<- fit$residuals # replace all data-cells with residual values resid <-predictors$NDVI values(resid) <-rval;rm(rval) names(resid) <- "Residuals"</pre> # Do the plot with spplot ss <- stack(s$Birds.richness, pred, resid) sp <- as(ss, 'SpatialGridDataFrame') trellis.par.set(sp.theme()) spplot(sp)
While looking at the residual plot you might notice that our simple model fails to explain all the variation at mountain altitudes (the Andes). Still the predicted values look very alike the observed richness. Bird species Richness is highest at tropical mountain ranges, which is consistent with results from Africa (Jetz & Rahbek, 2002). Reasons for this pattern are not fully understood yet, but if i had to discuss this with a colleague i would probably bring up arguments like older evolutionary time, higher habitat heterogeneity and greater numbers of climatic niches at mountain ranges. At this point you would then test for spatial autocorrelation using Moran´s I, adjust your data to that and use more sophisticated methods like General Additive Models (GAMs) or Spatial Autoregressive Model (SARs) and account for the spatial autocorrelation. See Rahbek et al. (2007) for the actual study.
References:
- Jetz, W., & Rahbek, C. (2002). Geographic range size and determinants of avian species richness. Science, 297(5586), 1548-1551.
- Rahbek, C., Gotelli, N. J., Colwell, R. K., Entsminger, G. L., Rangel, T. F. L., & Graves, G. R. (2007). Predicting continental-scale patterns of bird species richness with spatially explicit models. Proceedings of the Royal Society B: Biological Sciences, 274(1607), 165-174.
Interesting Paper: Impact of Fragmentation on Plant-Frugivore networks redundancy
Jörg Albrecht, my former co-supervisor at the University of Marburg finally published his first results from his PhD. I was eagerly waiting for this publication as i also helped to raise a lot of data as a volunteer ornithologist while working on my bachelor thesis. Looking back i remember many nice beautiful moments  like sitting in a camouflaged tent early in the morning counting frugivorous birds in the very core zone of Bialowieza forest. Bisons, Mooses, wildcats, the sound of howling wolfs in the morning were among the nice experiences i took with me. It was definitely a nice period of my life and i congratulate Jörg for publishing this nice paper! Published in the renown Journal of Ecology you can now access the paper in early-view:
like sitting in a camouflaged tent early in the morning counting frugivorous birds in the very core zone of Bialowieza forest. Bisons, Mooses, wildcats, the sound of howling wolfs in the morning were among the nice experiences i took with me. It was definitely a nice period of my life and i congratulate Jörg for publishing this nice paper! Published in the renown Journal of Ecology you can now access the paper in early-view:
For those of you who want a little appetizer of what the paper is about. The paper itself incorporates information gained from many recently developed techniques for ecological network analysis to draw conclusions using general ecological theory (Optimal Foraging Theory). The optimal Foraging Theory developed by MacArthur (1955) hasn’t yet much appeared in the ecological network literature, which is a shame as it allows predictions about changing plant/animal specialization patterns in the light of habitat perturbations. Here is a little summary, but i still recommend to read the full paper as both methods and conclusions are quite sophisticated 🙂
- 2 years of recorded plant-frugivorous interactions in Europe’s last old-growth lowland forest (Białowieza, Eastern Poland)
- Hypothesis (Summarized from the 3 expectations in the paper): Increased competition at Forest edges (caused by logging) compared to the interior forest leads to higher, respectively lower, redundancy in plant-frugivore networks. (But better read the paper!)
- To fully understand the extent of this study you need to dive deep into the study design and purpose. It uses state of the art statistical network-analysis techniques to calculate network redundancy and interaction specialization. Two and a half pages alone explain the data gathering and analysis, while the results sections is nearly half a page long 🙂
- Despite the small sample size (which was/is a major critical point) the results show that the networks redundancy was reduced at forest edges due to shifting dietary specialization of the interacting partners. As i remember from the data collection Black-caps and Blackbirds dominated in most of the assessed trees, which leads to an asymmetry in the interaction network. As shown in the paper this might be due to forest fragmentation.
- Possible Critic:The sampling effort of 10 studysites, which were sampled over 2 years might not be enough to detect real properties of ecological networks. However so far every study investigating ecological networks worked with incomplete data. I also don’t really trust the whole separation of species in specialist and generalists based on available literature. Although much is known up to now, both foraging and behavioral patterns of birds might change due to forest fragmentation and therefore such classification might be inaccurate depending on the study sites properties.
- Why is the study of interest? Up to now not much is known about how temperate Plant-frugivore interactions change in the face of habitat perturbation. Most of the available literature was conducted in the tropics or didn’t incorporated whole interaction networks. Probably due to the geographical bias, most of the available literature furthermore failed to detect an effect on network stability as this study shows.
So, enough advertising 😉
Calculating average percentual Home-range overlaps in R
If you are studying ornithology you can usually fall back on a rich basis of knowledge accumulated over many years by volunteers, birders and professional ornithologist. I can think of no other taxonomic group that has been studied this intensively (well, maybe plants). Any yet it amazes me that stil
l we lack so much knowledge and data about why bird populations are declining and what the reasons for these declines are. For instance since some years it has been known that especially long-distant migrants are quite vulnerable to multiple reasons like habitat degradation and climate change in their wintering grounds (Berthold et al. 1998, Sanderson et al. 2006; Heldbjerg & Fox 2008). Since so little is known (if for example compared to the American migration route) about how those species behave, forage and migrate in the areas below the Sahara, there is an urgent need for more information and data. Using the newest technology such as very tiny GPS loggers we became able to track many declining birds to their wintering grounds.
Right know i am working with relocation data from 3 warbler species, which is a quite a new way of thinking for me (waypoints instead of occurence points) and right now i am diving into the topic by analyzing this data to for instance estimate the amount of overlaps between many species.
In this R-code i show you how you can easily use the package adehabitat for R to estimate the home-range using an adhoc Kernel. In my example i have used a point-layer containing relocation data with a Date and Time and a Species ID (Bird_ID) of 3 warbler species in an area in southern Senegal.
require(adehabitat);library(rgdal)
# Read in your Relocation Layer
spp <- readOGR("source","layer")
# Construct an adhoc Kernel for my 3 different warbler species
k <- kernelUD(as.data.frame(coordinates(spp)),
id=spp$Bird_ID,h="href",same4all=T,grid=200)
# Calculate the area in square kilometers for a 50% Core-Zone and 95% Zone
kernel.area(data.frame(coordinates(spp)),
id=spp$Bird_ID,levels=c(50,95),unin="m",unout="km2")
# Calculate the Overlap of the kernel in Percent for all Bird species
(kov <- kerneloverlaphr(k,method="HR",lev=95)) # Percentage Overlap 95
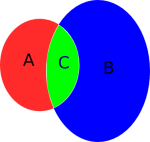 Calculating Percentage values of overlaps usually returns a matrix containing two different values. The percentage overlap from the smaller home-range to the bigger and vice versa (See Picture – Area C). Therefore there could be two different percentage values depending from which site you’re looking at it.
Calculating Percentage values of overlaps usually returns a matrix containing two different values. The percentage overlap from the smaller home-range to the bigger and vice versa (See Picture – Area C). Therefore there could be two different percentage values depending from which site you’re looking at it.
My approach for this problem has been to simply swap the distance matrix and average the percentage values (this is kinda inaccurate, but still used in some newer papers), but i also would advice to use other indices, like the Utilization distribution Overlap Index (UDOI) proposed by Fieberg and Kochanny in 2005.
# SwapMatrix function to convert a distance matrix into a data.frame
swapMatrix <- function(m) {
ut <- upper.tri(m)
data.frame(i = rownames(m)[row(m)[ut]],
j = rownames(m)[col(m)[ut]],
x=t(m)[ut],
y=(m)[ut])
}
m <- swapMatrix(kov) # Swap your overlap
m$avg <- rowMeans(m[,3:4]) # And calculate the average of both values
Thats be it for now. I am still in the process of exploring all those techniques to investigate species home-ranges. Any links or cool examples are much appreciated.
References:
- Berthold, P, Fiedler, W, Schlenker, R, Querner, U, “25-year study of the population development of Central European songbirds: a general decline, most evident in long-distance migrants.” Naturwissenschaften 85.7 (1998): 350-353.
- Sanderson, Fiona J, Donald, Paul F., Pain, Deborah J., Burfield, Ian J., van Bommel, Frans P.J. “Long-term population declines in Afro-Palearctic migrant birds.” Biological Conservation 131.1 (2006): 93-105.
- Heldbjerg, Henning, and Fox, Tony (AD) “Long-term population declines in Danish trans-Saharan migrant birds: Capsule Long-distance migrant birds show less favourable trends than sedentary/short-distance species.” Bird Study 55.3 (2008): 267-279.
- Fieberg, J. and Kochanny, C.O. (2005) Quantifying home-range overlap: the importance of the utilization distribution. Journal of Wildlife Management, 69, 1346–1359.
Studies, Copenhagen and Bird Lookup
Hej dear Readers,
sorry for the long absence, but i had and still have a lot of stuff going on. In pursue of a Master degree i recently moved to Copenhagen, Denmark. This comes with a lot of new obstacles and experiences for me. The focus of new blog posts will move slightly into the direction of my study focus and therefore you can expect some posts about phylogeny, ornithology or more theoretical stuff in the future.
By the way: There are still a lot of visits and blog comments about problems with my LecoS plugin. Please use this Bug-tracker for reporting future issues. I will try to fix them, if i have time and sudden coding passion.
As a little extra to this blog post:
I just cleaned my computer from all old stuff, raster data and other files. While doing this i stumbled upon a small little tool i coded back in 2005. It´s a simple bird-names lookup tool (called Birdlooker) coded in python. It takes a German, Latin or English bird name of the Western Palaearctic bird world as input and returns their respective translated names in the other languages. I coded it because i wanted a fast lookup of Latin and English names for German birds without using the Internet or a book.
Maybe it is still of use for some people. Download it here